Introduction. The role (and control) of fibroblast cell populations within the developing lung is not well understood. Here we explore PDGFRa+ fibroblast cells during lung development using PDGFRa+ GFP transgenic mice, with fluorescence imaging, RNA profiling & FACs analysis. From these analyses we are beginning to characterize specific cell populations and their interactions through development.
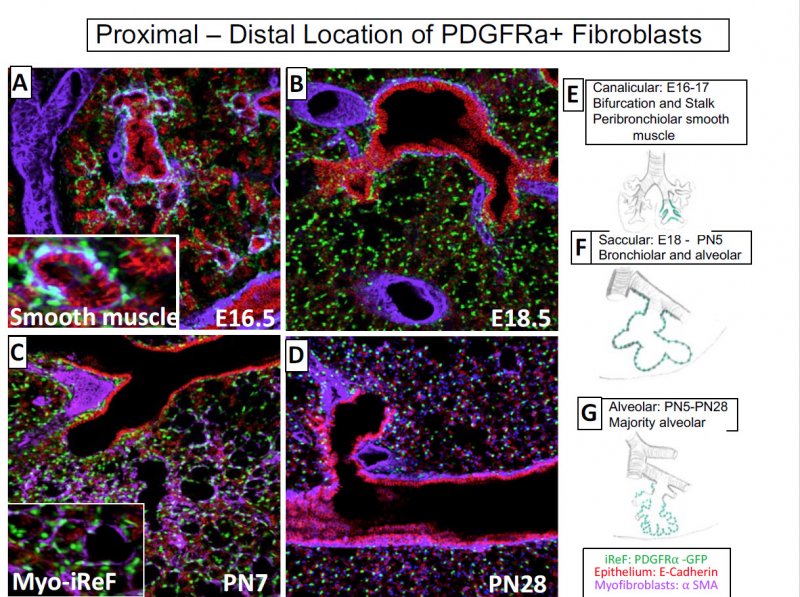
In this series of images from ages E16.5 to PN28, the nuclei of the fibroblasts of interest are stained in green using PDGFRa (platelet derived growth factor receptor, alpha) GFP expressing transgenic mice. Through the series of panels shown, you can see the spatial location of the staining fibroblasts in the images. In panel A, at age 16.5, you find PDGFRa expressed in the smooth muscle, particularly the bronchiolar stalks. At age 18.5, expression is lost around the bronachiolar stalks and seen among the periphery. By PN7 the cells in the periphery are myofibroblasts, but by PN28 the cells are no longer myofibroblasts. The cartoon diagrams (E-G) demonstrate the change in location of the cells through development, from proximal to distal to the alveolar region.
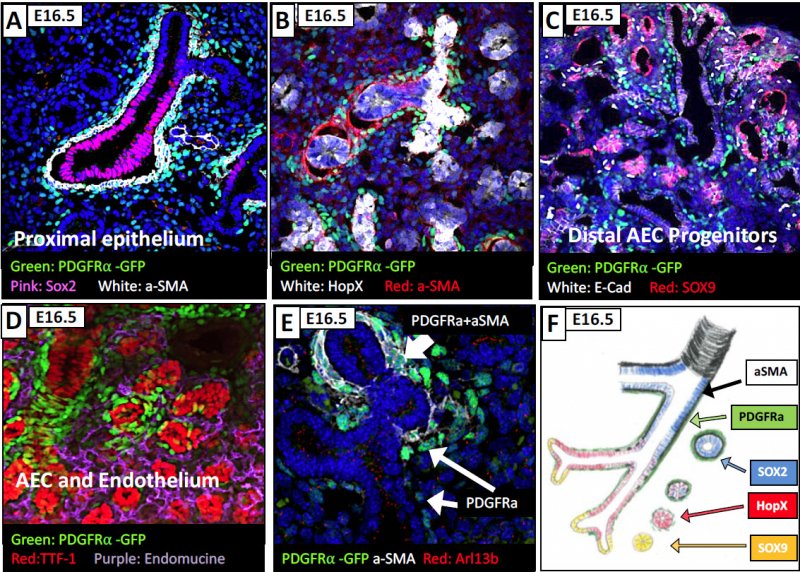
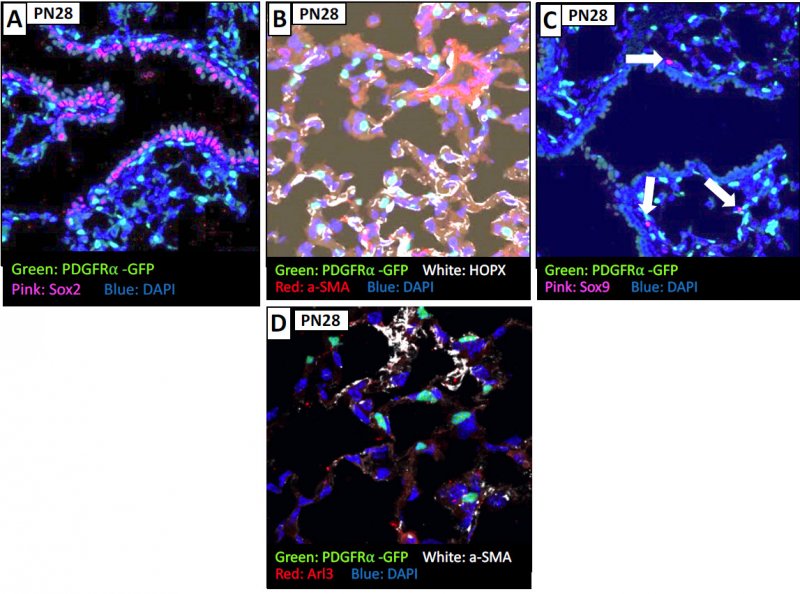
To explore what epithelial cells the fibroblasts "talk to", a series of markers were used to explore the proximal epithelium of the age 16.5 mouse: Sox2 marker (for upper bronchiolar cells), HOPX ( for transient bronchioles) , Sox9 (for terminal bronchioles). In panel A, the PDGFRa cells are found outside of the bronchiolar cells and some are SMA (smooth muscle actin) positive (white). In B, with the HOPX staining for the transient bronchioles, you see some PDGFRa+ cells that are SMA positive and some that are not. In Panel C, focusing in on the Sox9 region (terminal brochioles) the cells are absent from that region. D and E show some aditional cells for example, and F gives you the schematic of the spatial interpretation of the PDGFRa positive cells along the developing lung.
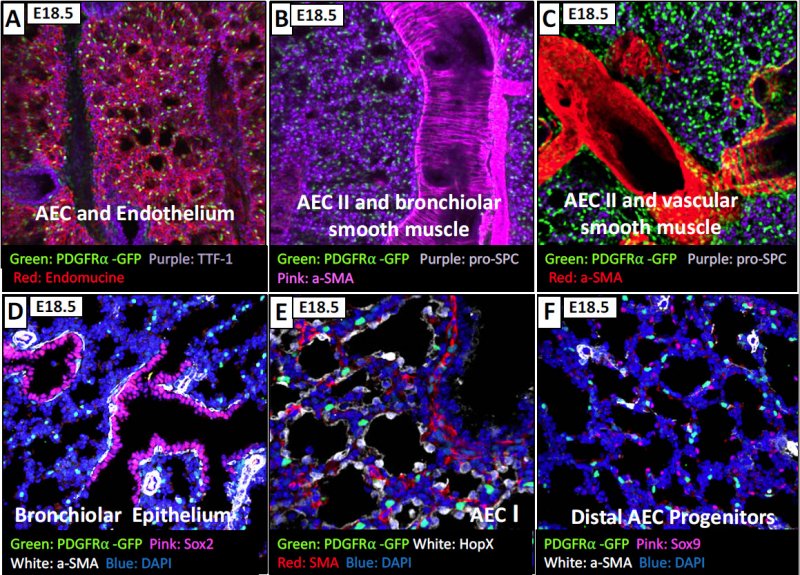
At age 18.5 you lose proximity to SOX2 epithelial expressing cells, and you find that the PDGFRa+ fibroblasts are not associated with Type I (AEC - alveolar epithelial cells) or type II (AEC) cells as well as the SOX9 (AEC progenitor) cells. Therefore, at this time we do not know what epithelial cells the PDGFRA+ fibroblasts are primarily associated with.
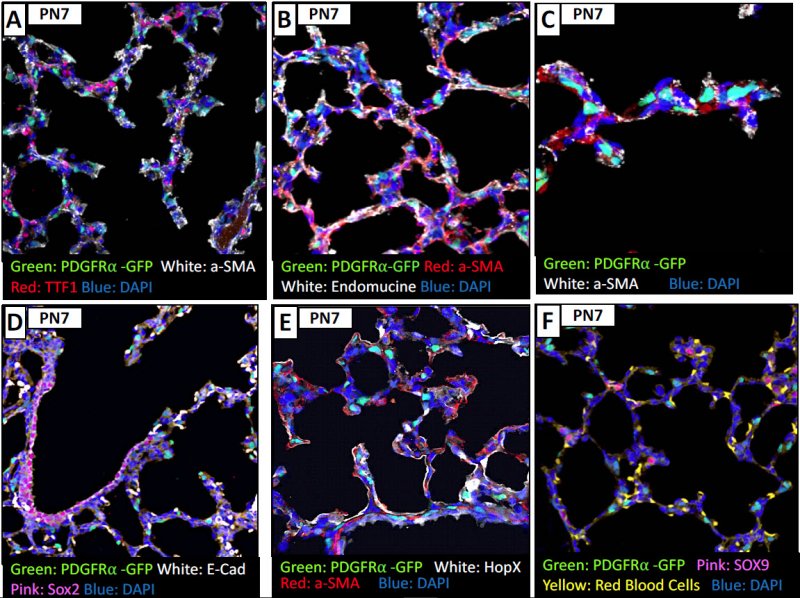
By age PN7 and PN28, the PDGFRa expressing cells are also expressing SMA, and some that are extruded. While it was initially thought that cells needed SMA to exclude, maybe is in relation to the forces in the region that causes the extrusion of the cells. Again, you see that the PDGRa+ cells are not colocalized with HOPx region or SOX9 regions at this time of development.
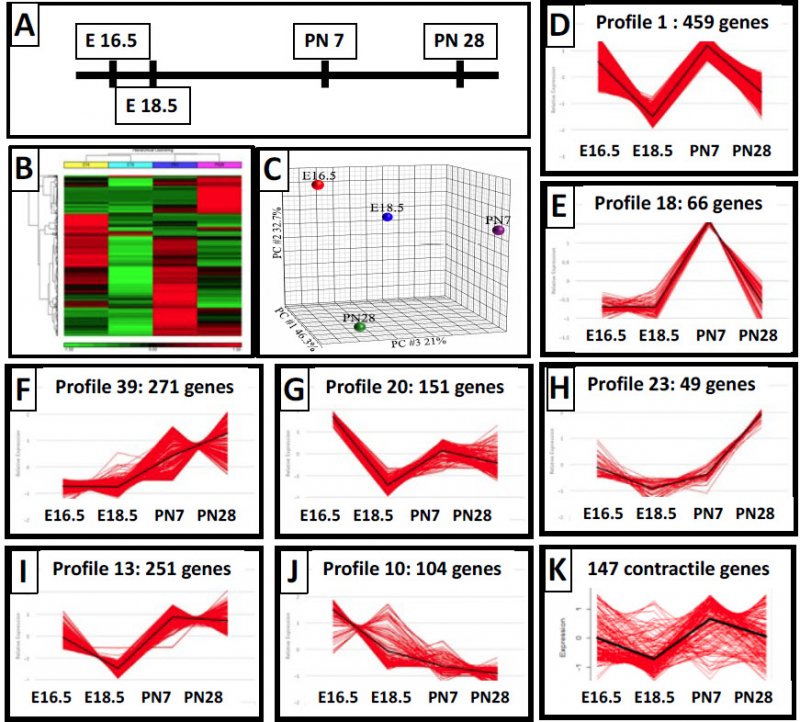
The PDGFRa+ cells change spatial location. Many express SMA and are myofibroblasts at the 16.5 and post natal timepoints. To further explore what is occurring with these myofibroblasts, we isolated RNA from fractionated cells to identify differentially expressed genes through development. About 2000 genes were identified (Panel B), and Panel C shows the PCN analysis across the developmental timepoints for the genes. We further profiled 500 of the genes that are co-regulated, but the categories were confusing for the genes. So we then clustered the genes based upon their expression pattern and looked at 7 such clusters and those genes within the clusters and came up with some interesting observations based upon the clusters (E-K).
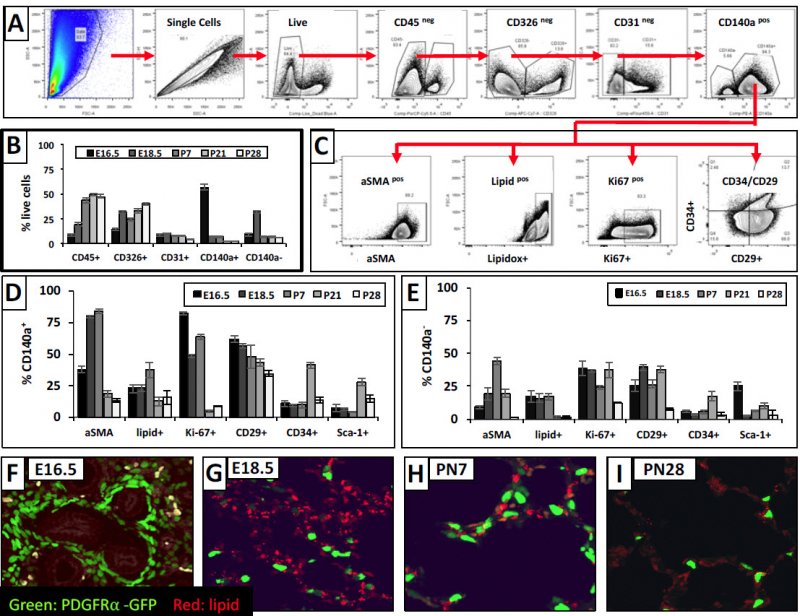
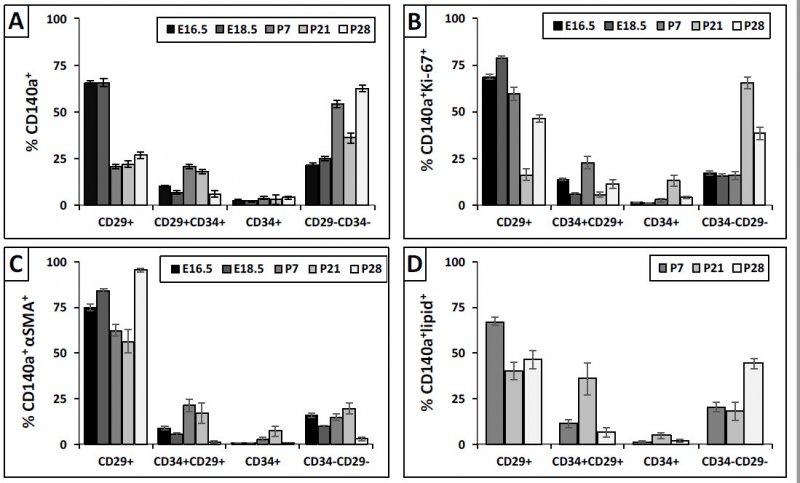
We used FACs analysis to further explore proliferation, SMA and lipids in the population of cells across the developmental timepoints further subdividing into CD34/CD29 cells. For the CD140+ (i.e., PDGFRA+) cells, you do find SMA is expressed at 18.5 and PN7 (Panel D) & not thereafter. Lipids peak at PN7, and proliferation peaks early on and tapers off at the later time points. Panels D-E provide the other markers that we analyzed in this experiment. Panels F-I show the lipid staining pattern, & we see lipid co-localization at P28, but not the earlier timepoints.
We analyzed further the PDGFRa+ CD29/CD34 subdivided fibroblasts for which one are expressing lipids, proliferating, or expressing SMA. You see (Panel C) that it is the CD29+ that are myofibroblasts, and double negatives at PN28 are the lipofibroblasts
Future endeavors: We are isolating the cells and sending them to other LungMAP laboratories to look at epigenetics and proteomics & lipidomics. We are also adding additional timepoints inbetween.