“The Lung---Cell by Cell” series is drawn from the publication “Building and Regenerating the Lung Cell by Cell” which compiles formative information derived from LungMAP experiments conducted by researchers at Cincinnati Children’s Hospital Medical Center led by Dr. Jeffrey Whitsett.
Cell data used in this paper were generated through single cell experiments. Lung samples were digested and single cells were purified by FACS (attachment), microfluidics or in microdroplets (DropSeq) and RNA was prepared for next generation sequencing.
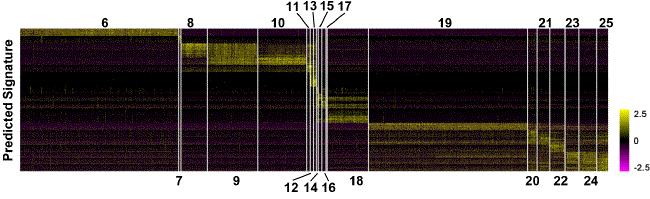
Cell sequencing output was interrogated using iterative hierarchical and graph-based clustering strategies----cells were statistically subclassified into major cell types and sub-types. In lung samples from post-natal day 1 mouse, the tSNE algorithm (dimensionality reduction similar to Principal Component Analysis) was used to perform clustering of Drop-Seq data, to yield four major cell types: Endothelial, Mesenchymal, Epithelial and Immune (left panel) and 25 subtypes (right panel).
Based on these defined cell tpes, binomial and negative probability testing was performed to identify differentially expressed genes and predict signature gene characteristics for each cell type shown above. Prediction results are shown as a heatmap below.
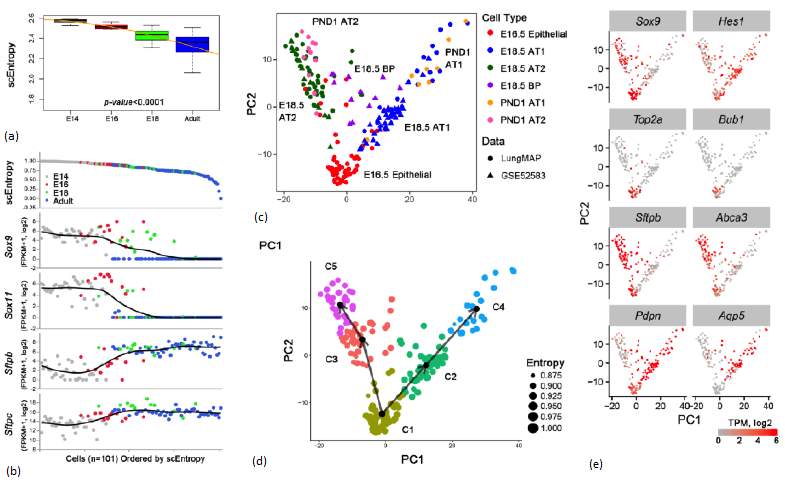
Cell types are readily clustered on the basis of RNA profiles that, together with antibody, lineage tracing, and in situ hybridization, validate cell type specificity provided by the transcriptomic data. Since multiple cell types interact and differentiate during organogenesis, distinct cell "states" of differentiation, proliferation, and cellular response are evident, even within a seemingly homogeneous populations of cells. Sequential relatedness of cells can be predicted statistically in "pseudotime" or ordered by single cell entropy.
Single cell entropy (scEntropy) is a method to measure differentiation states of individual cells, thus it can be used to predict sequential relatedness of cells. Here we show a model predicting the lineage trajectory of epithelial progenitor cells during mouse lung development using SLICE, an algorithm developed to determine cell differentiation and lineage based on single cell entropy (109). At E16.5 of mouse lung gestation, many epithelial cells are highly proliferative and remain relatively poorly differentiated.